Box 6.1. Molecular insights into insect development
The formation of segments in the early embryo of Drosophila is understood better than almost any other complex developmental process. Segmentation is controlled by a hierarchy of proteins known as transcription factors, which bind to DNA and act to enhance or repress the production of specific messages. In the absence of a message, the protein for which it codes is not produced; thus ultimately transcription factors act as molecular switches, turning on and off the production of specific proteins. In addition to controlling genes below them in the hierarchy, many transcription factors also act on other genes at the same level, as well as regulating their own concentrations. Mechanisms and processes observed in Drosophila have much wider relevance, including to vertebrate development, and information obtained from Drosophila has provided the key to cloning many human genes. However, we know Drosophila to be a highly derived fly, and it may not be a suitable model from which to derive generalities about insect development.
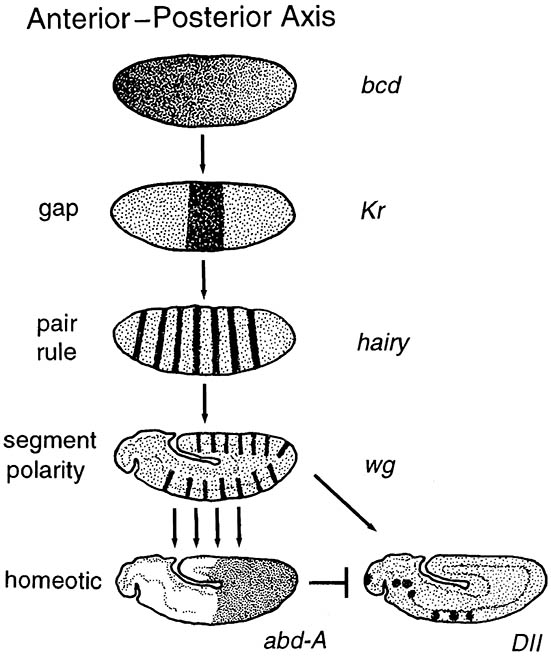
During oogenesis (section 6.2.1) in Drosophila, the anterior—posterior and dorsal—ventral axes are established by localization of maternal messenger RNAs (mRNAs) or proteins at specific positions within the egg. For example, the mRNAs from the bicoid (bcd) and nanos genes become localized at anterior and posterior ends of the egg, respectively. At oviposition, these messages are translated and proteins are produced that establish concentration gradients by diffusion from each end of the egg. These protein gradients differentially activate or inhibit zygotic genes lower in the segmentation hierarchy — as in the upper figure (after Nagy 1998), with zygotic gene hierarchy on the left and representative genes on the right — as a result of their differential thresholds of action. The first class of zygotic genes to be activated is the gap genes, for example Kruppel (Kr), which divide the embryo into broad, slightly overlapping zones from anterior to posterior. The maternal and gap proteins establish a complex of overlapping protein gradients that provide a chemical framework that controls the periodic (alternate segmental) expression of the pair-rule genes. For example, the pair-rule protein hairy is expressed in seven stripes along the length of the embryo while it is still in the syncytial stage. The pair-rule proteins, in addition to the proteins produced by genes higher in the hierarchy, then act to regulate the segment polarity genes, which are expressed with segmental periodicity and represent the final step in the determination of segmentation. Because there are many members of the various classes of segmentation genes, each row of cells in the anterior—posterior axis must contain a unique combination and concentration of the transcription factors that inform cells of their position along the anterior—posterior axis.
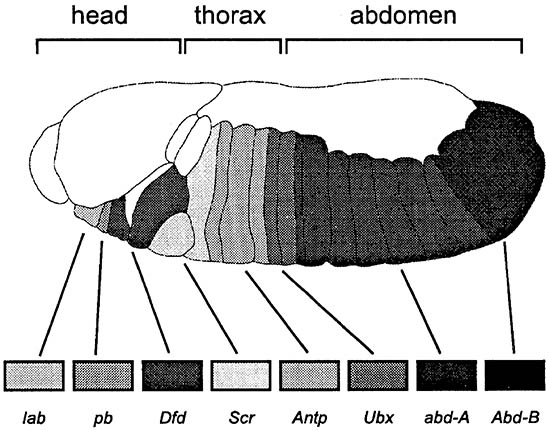
Once the segmentation process is complete each developing segment is given its unique identity by the homeotic genes. Although these genes were first discovered in Drosophila it has since been established that they are very ancient, and a more or less complete subset of them is found in all multicellular animals. When this was realized it was agreed that this group of genes would be called the Hox genes, although both terms, homeotic and Hox, are still in use for the same group of genes. In many organisms these genes form a single cluster on one chromosome, although in Drosophila they are organized into two clusters, an anteriorly expressed Antennapedia complex (Antp-C) and a posteriorly expressed Bithorax complex (Bx-C). The composition of these clusters in Drosophila is as follows (from anterior to posterior): (Antp-C) — labial (lab), proboscidea (pb), Deformed (Dfd), Sex combs reduced (Scr), Antennapedia (Antp); (Bx-C) — Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B), as illustrated in the lower figure of a Drosophila embryo (after Carroll 1995; Purugganan 1998). The evolutionary conservation of the Hox genes is remarkable for not only are they conserved in their primary structure but they follow the same order on the chromosome, and their temporal order of expression and anterior border of expression along the body correspond to their chromosomal position. In the lower figure the anterior zone of expression of each gene and the zone of strongest expression is shown (for each gene there is a zone of weaker expression posteriorly); as each gene switches on, protein production from the gene anterior to it is repressed.
The zone of expression of a particular Hox gene may be morphologically very different in different organisms so it is evident that Hox gene activities demarcate relative positions but not particular morphological structures. A single Hox gene may regulate directly or indirectly many targets; for example, Ultrabithorax regulates some 85–170 genes. These downstream genes may operate at different times and also have multiple effects (pleiotropy); for example, wingless in Drosophila is involved successively in segmentation (embryo), Malpighian tubule formation (larva), and leg and wing development (larva—pupa).
Boundaries of transcription factor expression are important locations for the development of distinct morphological structures, such as limbs, tracheae, and salivary glands. Studies of the development of legs and wings have revealed something about the processes involved. Limbs arise at the intersection between expression of wingless, engrailed, and decapentaplegic (dpp), a protein that helps to inform cells of their position in the dorsal—ventral axis. Under the influence of the unique mosaic of gradients created by these gene products, limb primordial cells are stimulated to express the gene distal-less (Dll) required for proximodistal limb growth. As potential limb primordial cells (anlage) are present on all segments, as are limb-inducing protein gradients, prevention of limb growth on inappropriate segments (i.e. the Drosophila abdomen) must involve repression of Dll expression on such segments. In Lepidoptera, in which larval prolegs typically are found on the third to sixth abdominal segments, homeotic gene expression is fundamentally similar to that of Drosophila. In the early lepidopteran embryo Dll and Antp are expressed in the thorax, as in Drosophila , with abd-A expression dominant in abdominal segments including 3–6, which are prospective for proleg development. Then a dramatic change occurs, with abd-A protein repressed in the abdominal proleg cell anlagen, followed by activation of Dll and up-regulation of Antp expression as the anlagen enlarge. Two genes of the Bithorax complex (Bx-C), Ubx and abd-A, repress Dll expression (and hence prevent limb formation) in the abdomen of Drosophila. Therefore, expression of prolegs in the caterpillar abdomen results from repression of Bx-C proteins thus derepressing Dll and Antp and thereby permitting their expression in selected target cells with the result that prolegs develop.
A somewhat similar condition exists with respect to wings, in that the default condition is presence on all thoracic and abdominal segments with Hox gene repression reducing the number from this default condition. In the prothorax, the homeotic gene Scr has been shown to repress wing development. Other effects of Scr expression in the posterior head, labial segment, and prothorax appear homologous across many insects, including ventral migration and fusion of the labial lobes, specification of labial palps, and development of sex combs on male prothoracic legs. Experimental mutational damage to Scr expression leads, amongst other deformities, to appearance of wing primordia from a group of cells located just dorsal to the prothoracic leg base. These mutant prothoracic wing anlagen are situated very close to the site predicted by Kukalová-Peck from paleontological evidence (section 8.4, Fig. 8.4b). Furthermore, the apparent default condition (lack of repression of wing expression) would produce an insect resembling the hypothesized “protopterygote”, with winglets present on all segments.
Regarding the variations in wing expression seen in the pterygotes, Ubx activity differs in Drosophila between the meso- and metathoracic imaginal discs; the anterior produces a wing, the posterior a haltere. Ubx is unexpressed in the wing (mesothoracic) imaginal disc but is strongly expressed in the metathoracic disc, where its activity suppresses wing and enhances haltere formation. However, in some studied nondipterans Ubx is expressed as in Drosophila — not in the fore-wing but strongly in the hind-wing imaginal disc — despite the elaboration of a complete hind wing as in butterflies or beetles. Thus, very different wing morphologies seem to result from variation in “down- stream” response to wing-pattern genes regulated by Ubx rather than from homeotic control.
Clearly, much is yet to be learnt concerning the multiplicity of morphological outcomes from the interaction between Hox genes and their downstream interactions with a wide range of genes. It is tempting to relate major variation in Hox pathways with morphological disparities associated with high-level taxonomic rank (e.g. animal classes), more subtle changes in Hox regulation with intermediate taxonomic levels (e.g. orders/suborders), and changes in downstream regulatory/functional genes perhaps with suborder/ family rank. Notwithstanding some progress in the case of the Strepsiptera (q.v.), such simplistic relationships between a few well-understood major developmental features and taxonomic radiations may not lead to great insight into insect macroevolution in the immediate future. Estimated phylogenies from other sources of data will be necessary to help interpret the evolutionary significance of homeotic changes for some time to come.
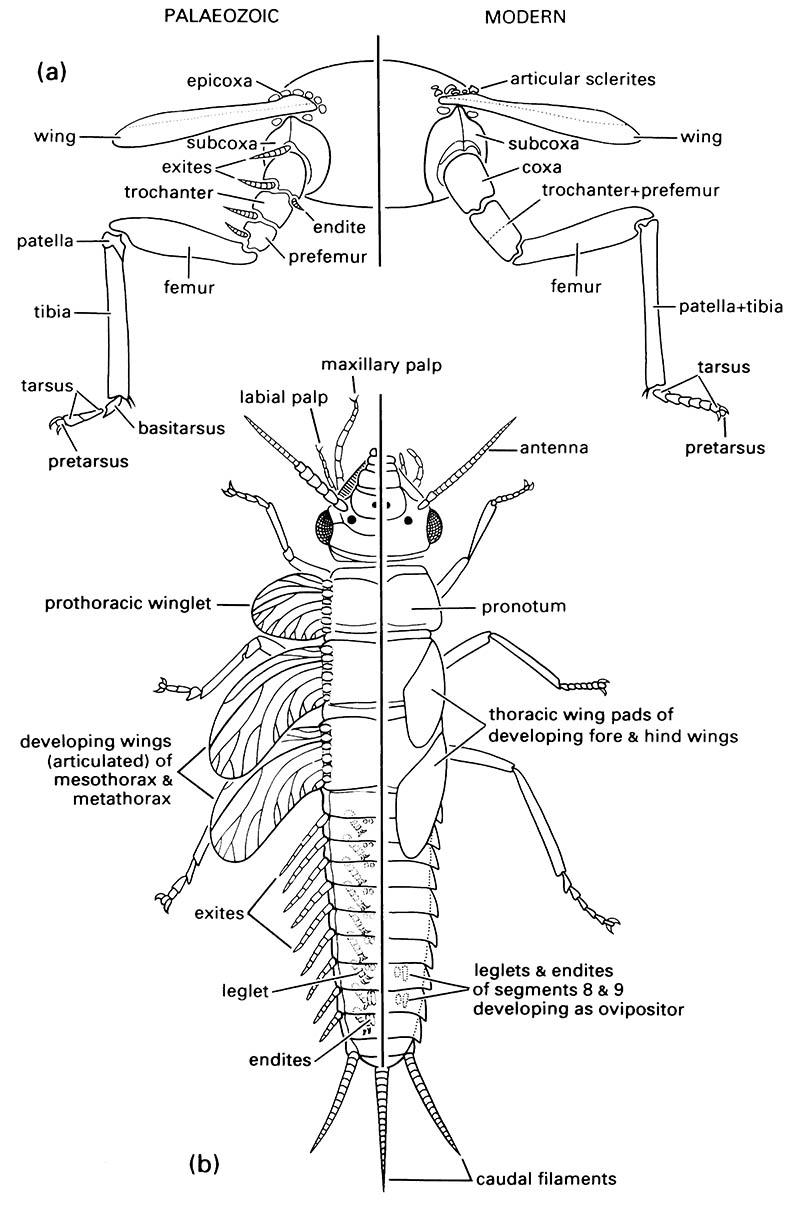
(a) thoracic segment of adult showing generalized condition of appendages; (b) dorsal view of nymphal morphology. (Modified from Kukalová-Peck 1991; to incorporate ideas of J.W.H. Trueman (unpublished))



